News
Exploring uncharted territories in RAS signaling leads to many surprises
Published on October 14, 2020
There are over 160 small GTPase proteins in human cells that can fluctuate between an active state (“on”) and an inactive state (“off”). These work as switches to control whether intracellular signaling pathways are turned on or off. Of particular interest are GTPases of the RAS family. Mutations in these GTPases are among the main causes of human cancers, yet there are currently no therapeutics targeting RAS proteins themselves.
Acting together with RAS are ‘effector’ proteins, which specifically recognize and complex with activated RAS to relay signals downstream, supporting the activity of signaling pathways that stimulate processes such as cell proliferation. These effector proteins are attractive therapeutic targets, and a better understanding of their functions could have a broad impact on the diagnosis, treatment and remission of cancer patients.
The RASSF proteins, intriguing but poorly studied
The group of Professor Matthew Smith at IRIC, and their collaborators at the Hebrew University of Jerusalem, undertook a systematic analysis of the poorly studied members of the RAS-association domain family (RASSF) effectors. Their surprising findings were just published in the journal Science Signaling. RASSF proteins are highly intriguing since they are thought to act as tumor suppressors by directly linking the RAS pathway with at least one other important signalling pathway, the Hippo pathway, to induce cell death instead of proliferation. Yet, the specific role of the 10 proteins of this family in signalling downstream of RAS and whether all 10 are true RAS effectors remained open questions until now.
Many surprises
The researchers first generated purified forms of these RASSF proteins as a toolbox to assay their interactions with activated RAS GTPases. To their surprise, only one of ten family members, RASSF5, binds activated RAS. In further contrast with previous studies, they found that no RASSFs stimulate programmed cell death (apoptosis) downstream of RAS oncoproteins. Interestingly, RASSF1-6 associates with a mammalian Hippo kinase and the other RASSFs interact with regulators of p53, an important tumor suppressor with a pro-apoptotic function.
New binding partners
Having discovered that most RASSFs do not interact with the RAS GTPase oncoproteins, the researchers then measured directly their interactions with related GTPases previously proposed to bind RASSFs. They could not corroborate a single interaction between RASSFs and any of these potential targets, suggesting the RAS-related GTPases, which signal through these effectors, are unknown. They constructed structural models for RASSFs to guide the selection of putative new binding partners. This structure and sequence-based informatics approach led to the identification of six novel GTPase partners (including GTPases of the RGK family) for the well-known tumour suppressor protein RASSF1. Importantly, interplay between RASSF1 and these RGK GTPases activated the apoptotic Hippo pathway in cells.
This systematic analysis of RASSF binding partners is a valuable first step in defining the molecular mechanisms through which these effectors couple different RAS subfamily members to the Hippo pathway. As Hippo signaling is an emergent characteristic of RAS-driven cancers and their resistance to therapies, these data will eventually facilitate development of new therapeutics to be used in conjunction with other inhibitors, towards permanently halting the growth and relapse of RAS-driven cancer cells.
Referenced study
Dhanaraman, T., S. Singh, R.C. Killoran, A. Singh, X. Xu, J. Shifman, M.J. Smith.
RASSF effectors couple diverse RAS subfamily GTPases to the Hippo pathway.
Science Signaling
https://stke.sciencemag.org/content/13/653/eabb4778
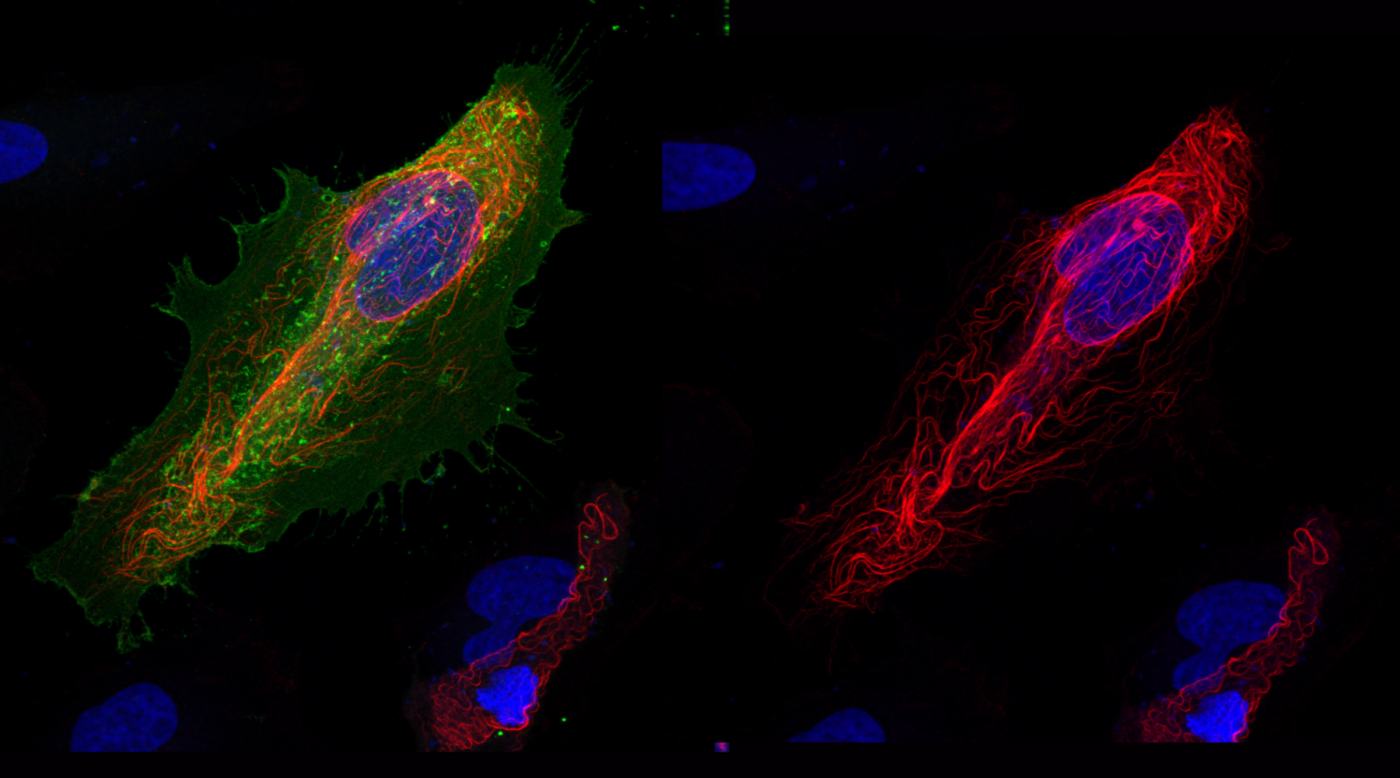
Photo legend
RAS subfamily GTPase binding to effector proteins is highly dynamic, tightly regulated and can significantly impact cellular physiology. In interphase HeLa cells RASSF1 resides on microtubules (red) while the RAS-related small GTPase REM1 (green) is localized at the plasma membrane.